New Features
Explore Ligand Quality
09/07
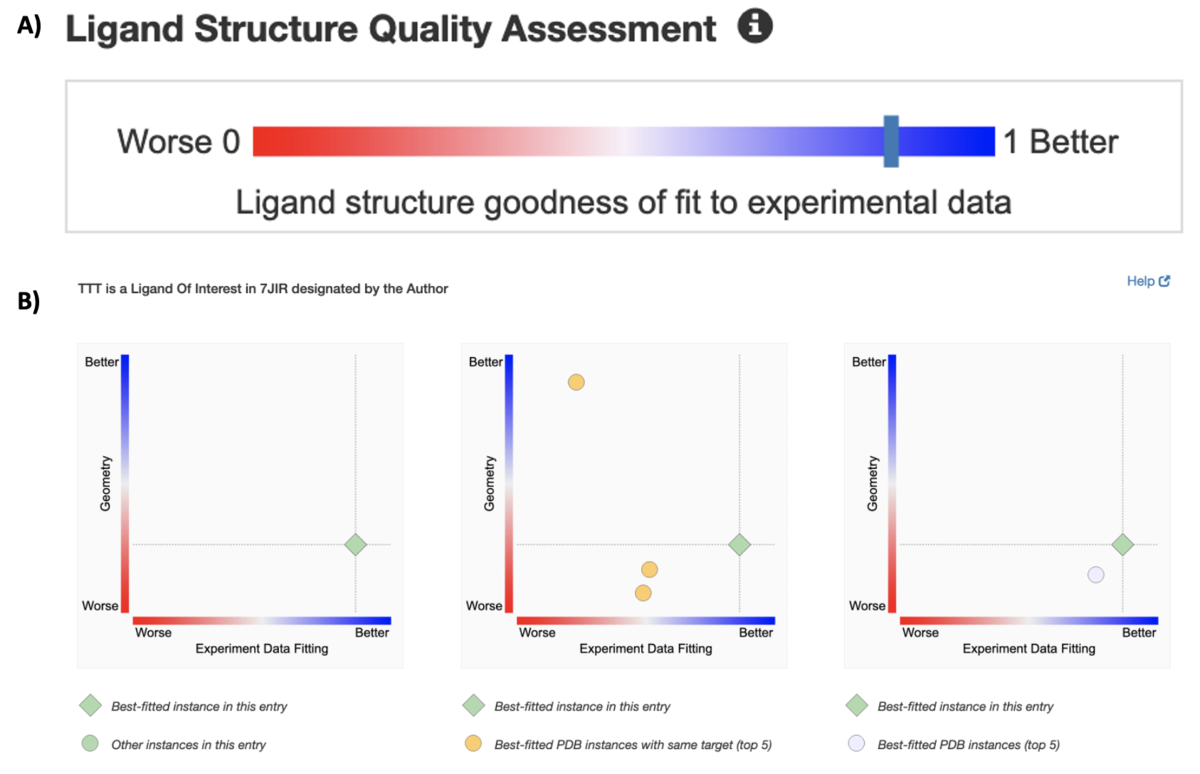 New graphics summarize ligand quality assessments. A) The slider graphic on the Structure Summary page for the PDB structure of a SARS-CoV-2 papain-like proteinase of 7JIR indicates the fitting quality of the ligand of interest. Clicking on the bar will take the user to the Ligands tab for more information about this ligand in this particular entry and across the archive.
New graphics summarize ligand quality assessments. A) The slider graphic on the Structure Summary page for the PDB structure of a SARS-CoV-2 papain-like proteinase of 7JIR indicates the fitting quality of the ligand of interest. Clicking on the bar will take the user to the Ligands tab for more information about this ligand in this particular entry and across the archive.More than 70% of PDB structures are complexed with small molecule ligands. These complexes represent a range of "quality" in terms of how well the atomic coordinates are supported by experimental data, and how well the ligand 3D structure agrees with known chemistry (e.g., bond lengths, bond angles).
New ligand structure quality assessment metrics are accessible from Structure Summary pages for PDB structures determined by X-ray crystallography. Correlated quality indicators (e.g., RSR, RSCC) have been aggregated into a ranking score that can be used for comparison across the archive.
Quality information for Ligand/s Of Interest is indicated by a slider bar on the Structure Summary page for a PDB structure (e.g., 7JIR). Clicking on the bar representing the ligand returns the Ligands tab, which provides more information about this ligand in this particular entry and for the top five "high quality" instances in the archive.
The new indicators allow any user to quickly review ligand structure quality and unambiguously select the CCD ligand (or ligands) in a particular PDB structure that will best serve their research or teaching interests.
Detailed documentation about this feature is available; for more information about small molecule ligand validation:
- Enhanced Validation of Small-Molecule Ligands and Carbohydrates in the Protein Data Bank (2021) Structure 29: 393-400.e391 doi: 10.1016/j.str.2021.02.004
- Outcome of the First wwPDB/CCDC/D3R Ligand Validation Workshop (2016) Structure 24:502-508 doi: 10.1016/j.str.2016.02.017












