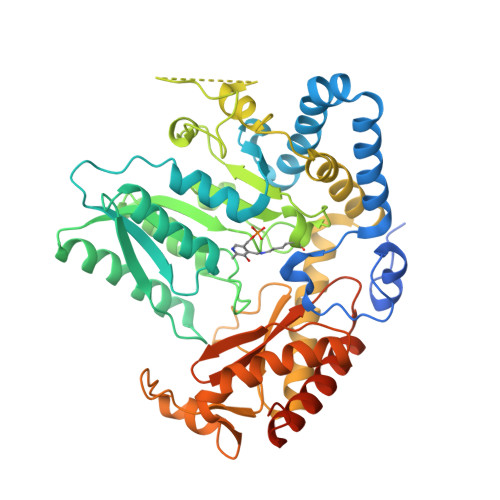
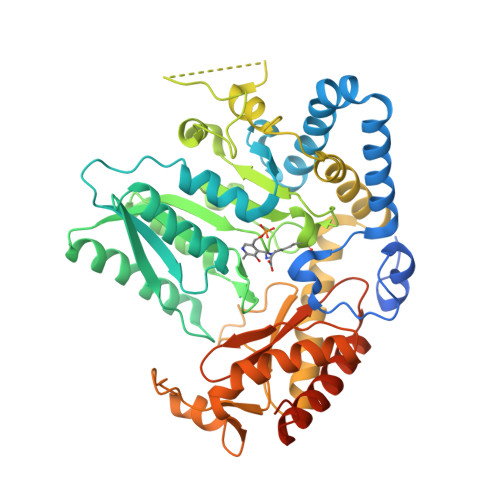
Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Irani, S., Naowarojna, N., Tang, Y., Kathuria, K.R., Wang, S., Dhembi, A., Lee, N., Yan, W., Lyu, H., Costello, C.E., Liu, P., Zhang, Y.J.(2018) Cell Chem Biol 25: 519-529.e4
- PubMed: 29503207
- DOI: https://doi.org/10.1016/j.chembiol.2018.02.002
- Primary Citation of Related Structures:
5UTS, 5V12, 5V1X - PubMed Abstract:
Sulfur incorporation in the biosynthesis of ergothioneine, a histidine thiol derivative, differs from other well-characterized transsulfurations. A combination of a mononuclear non-heme iron enzyme-catalyzed oxidative C-S bond formation and a subsequent pyridoxal 5'-phosphate (PLP)-mediated C-S lyase reaction leads to the net transfer of a sulfur atom from a cysteine to a histidine. In this study, we structurally and mechanistically characterized a PLP-dependent C-S lyase Egt2, which mediates the sulfoxide C-S bond cleavage in ergothioneine biosynthesis. A cation-π interaction between substrate and enzyme accounts for Egt2's preference of sulfoxide over thioether as a substrate. Using mutagenesis and structural biology, we captured three distinct states of the Egt2 C-S lyase reaction cycle, including a labile sulfenic intermediate captured in Egt2 crystals. Chemical trapping and high-resolution mass spectrometry were used to confirm the involvement of the sulfenic acid intermediate in Egt2 catalysis.
- Department of Chemical Engineering, University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: